For additional questions, please reach out to us.
General FAQ
How do I submit my samples and where do I ship them?
Prior to dropping off any samples, clients at the University of Guelph should place a request through BookitLab (https://core.bookitlab.com/aac/). Samples can be dropped off at the genomics facility in room 1401 of the Summerlee Science Complex where you can print tube and plate labels for your submission.
External Clients
Clients outside of the University of Guelph can ship to:
University of Guelph
Advanced Analysis Centre - Genomics Facility
488 Gordon Street
Summerlee Science Complex (Bld 140)
Room 1401
Guelph, ON, Canada
N1G 2W1
Where can I get a quote?
Please contact us for placing customized orders.
How can I book equipment?
Can you do custom library preparation?
Yes! Please contact us for details and to generate a quote.
How do I download my data?
For large downloads, you will be provided with a download link to use with our download manager (available for Windows, Linux, and Macintosh). Data will remain accessible for one month unless arranged otherwise. Alternate forms of data delivery, such as SFTP, are available but may incur additional fees. Quality control reports will be sent by email. Sanger sequencing and fragment analysis results will be sent by email.
Next Generation Sequencing
How should I provide my samples for NGS?
| # of Samples | Recommended storage tube/plate |
|---|---|
| 1 to 8 | Individual tubes |
| 8 to 24 | 8 tube strips or part of a 96 well PCR plate* |
| More than 24 | Part of whole 96 well PCR plate* |
*Organize samples vertically (A1-H1, A2-H2) — NOT horizontally (A1-A12).
What NGS applications do you support?
We can support many common NGS protocols from genome sequencing to transcriptomics. Long read and single cell sequencing are also available. We often are able to accommodate custom library preps or specific kits not listed below. If you have any questions about whether we can support your sequencing project please contact us.
Illumina Sequencing:
- Whole Genome/Shotgun Metagenome (Illumina DNA Prep, Illumina DNA Prep PCR-Free)
- RNA-Seq (NEB Ultra II RNA Prep, NEB UltraExpress RNA Kit)
- Amplicon Sequencing (16S/18S/ITS/custom)
- Single Cell (10X Genomics)
Oxford Nanopore:
PromethION 48 (Library preps TBD).
Can I submit sample libraries I prepared myself?
Yes! We do accept DIY libraries for Illumina sequencing, though we cannot guarantee library quality or success. Library quality control will be needed to ensure run integrity. Please contact us for more information.
What are your requirements for sample concentration and quantity?
| Application | Minimum Concentration | Minimum Quantity |
|---|---|---|
| 16S Sequencing | 5 ng/µL | 10 µL |
| Amplicon Sequencing | 5 ng/µL | 10 µL |
| Transcriptomics | — | — |
| Metagenomics | — | — |
| Whole genome sequencing | — | — |
Samples outside of the recommended concentrations can still be accepted but success is not guaranteed. Please contact us if this may be an issue for your project.
Which sequencing platforms do you have?
Currently we perform next generation sequencing on the Illumina NextSeq 1000. Soon, we will also support the Illumina NovaSeqX and PromethION 48
What sequencing depth do I need for my experiment?
Sequencing depth will vary considerably for different projects and often relates to the goals of the sequencing experiment. Please contact us to arrange a consultation if you are having trouble determining the appropriate amount of sequencing you require for your project.
Do you run QC on my samples?
We do not run QC on submitted samples unless requested. Please follow sample submission quality guidelines. After library preparation, we confirm library quality via fragment analysis on the Agilent TapeStation.
How long do you store unused sample and library?
We store unused sample for one month. Additional storage is available on request.
How long is my data stored?
Data will be stored for at least 1 month. Backups may be available on request, but we do not guarantee data availability after 1 month.
What is your sequencing turnaround time?
Turnaround time is dependent on the current queue. Orders placed that consume an entire flow cell can often be processed sooner, but please reach out to us if you have questions about the current estimated wait time.
How can I ensure successful sequencing?
Good quality sequencing starts with good quality samples. Make sure you are using validated extraction methods and always run QC on your samples prior to submitting them to us. We always do our best to ensure the highest possible probability of success, but we cannot guarantee it.
What is your policy for failed sequencing samples?
If quality control indicates that a success is possible, failed sequencing will be run a second time with no additional charge, except in cases when batch effects would render the repeat useless.
Can you provide analysis?
Yes! We currently offer bioinformatic pipelines for differential expression analysis (transcriptomics), metagenomics, phylogenetic assignment (16S/18S/ITS), variant calling, de-novo assembly, GO annotation/enrichment, and more. Please contact us for consultation or for generating a quote.
Sanger Sequencing
What are your recommendations for DNA concentration and quantity?
| DNA Template Size | Concentration (ng/µl) | Quantity (ng/rxn) |
|---|---|---|
| 100–500 bp | 5–10 | 20 |
| 500–1000 bp | 10–30 | 50 |
| 1 kb–5 kb | 30–100 | 150 |
| 5 kb–10 kb | 100–200 | 300 |
Please provide 3-10 µl of sample. We will accept samples outside these guidelines, but sequencing success rate will be lessened.
Do you run QC on my samples?
We do not verify the quality nor concentration of submitted samples for Sanger sequencing. Every sequencing run (excluding DIY submissions) includes an in-house control—if quality thresholds are not met, sequencing will be redone at no cost to you. For other sequencing concerns please see our troubleshooting notes below.
How long do you store unused sample?
We store unused sample for one month. Additional storage is available on request.
How long is my data stored?
Data will be stored for at least 1 month. Backups may be available on request, but we do not guarantee data availability after 1 month.
What is your sequencing turnaround time?
The typical turnaround for Sanger sequencing is 48 hours.
What can cause failed sequencing?
Failed sequencing can be caused by incorrect template concentration, poor quality DNA, badly designed primers, contamination, or machine failure (e.g. blocked capillary). To troubleshoot poor sequencing, we recommend looking at your trace files. For instance consider the following examples:
 This sample features a large dye blob, a result of contaminating DNA or residual dNTPs in the sample causing a peak of fluorescent signal. In this example, the signal strength of the sample is weak, and therefore it is very difficult to determine the sequence in this region. This may indicate that not enough template was present in the BigDye reaction.
This sample features a large dye blob, a result of contaminating DNA or residual dNTPs in the sample causing a peak of fluorescent signal. In this example, the signal strength of the sample is weak, and therefore it is very difficult to determine the sequence in this region. This may indicate that not enough template was present in the BigDye reaction.
 In this example, a small dye blob is still present in the same region of the spectrum, but since signal strength is much higher bases can still be accurately called in the region. If you are having problems with signal strength make sure you have accurately determined the concentration of your sample prior to submission, and that your primers are targeting the region of interest as expected.
In this example, a small dye blob is still present in the same region of the spectrum, but since signal strength is much higher bases can still be accurately called in the region. If you are having problems with signal strength make sure you have accurately determined the concentration of your sample prior to submission, and that your primers are targeting the region of interest as expected.
 In this example, many peaks can be seen interleaved on top of one another. This typically results from a mixed sample, if you had prepared amplicons from a taxon of interest it is possible that your sample was contaminated. It may also represent off target effects of your primers.
In this example, many peaks can be seen interleaved on top of one another. This typically results from a mixed sample, if you had prepared amplicons from a taxon of interest it is possible that your sample was contaminated. It may also represent off target effects of your primers.
For further assistance with Sanger sequencing troubleshooting, please reach out to us for a consultation.
What is your policy for failed sequencing samples?
Every sequencing run (excluding DIY submissions) includes an in-house control—if quality thresholds are not met, sequencing will be redone at no cost to you.
Can you design primers?
We can provide guidance, but do not design primers. Please reach out to us for a consultation.
Which primers do you have available?
A comprehensive list of primers offered by the facility can be found here.
Can you run Sanger sequencing with my own primers?
Yes! We are able to use any primer for the reaction. Please provide an aliquot along with your samples containing a 5 nM dilution of your primer with 2 uL per reaction using this primer. You can request multiple different primers with the same sequencing request.
How will I receive my data?
We provide both raw .ab1 and .seq files from the genetic analyzer via email. For other methods of transfer, please contact us.
Do you provide any bioinformatic analyses for Sanger sequencing?
Please contact us for a consultation for any further sample processing.
Gene expression services (PCR)
How much DNA do I need?
This can vary depending on application and scope of the project. Please reach out to us for a consultation.
What dye do you use for RT-PCR?
We use SsoAdvanced Universal Inhibitor-Tolerant SYBR Green Supermix which contains SYBR Green I dye for our RT-PCR reactions.
How does digital droplet PCR work, and what makes it different from RT-PCR?
In RT-PCR, a single sample is measured over time, but in Droplet Digital PCR, the sample is partitioned into 20,000 nanoliter-sized droplets. ddPCR provides better precision (±10%) and is capable of detecting small fold-changes in gene expression (1.2x-fold).
Should I use RT-PCR or ddPCR?
ddPCR is preferred for quantifying rare DNA targets, or in cases where a small difference between samples must be detected. Otherwise, traditional RT-qPCR can be used to save costs.
Can you provide primer design?
We can provide guidance, but do not design primers. Please reach out to us for a consultation.
DNA/RNA Quantification
What quantification methods do you have?
We offer both Qubit and NanoDrop for DNA and RNA quantification. Samples can be submitted to our lab for Qubit analysis.
We maintain a NanoDrop for client use based upon a subscription service (usually on a per lab group basis). The NanoDrop can be booked through BookitLab. For first time users please contact the facility to receive training.
Should I use the Qubit or the NanoDrop?
Qubit is more accurate and generally recommended for applications where a more precise concentration is needed such as NGS library preparation. NanoDrop can be useful for applications where a precise concentration is not necessarily required, or where absorbance ratios are desired (260/280 and 260/230).
What does good quality DNA/RNA on the NanoDrop look like?
When using NanoDrop, it is important to look at more than just your concentration. Contaminants in your sample can throw off accurate quantification. The image below shows an example of what a good curve on the NanoDrop should look like.
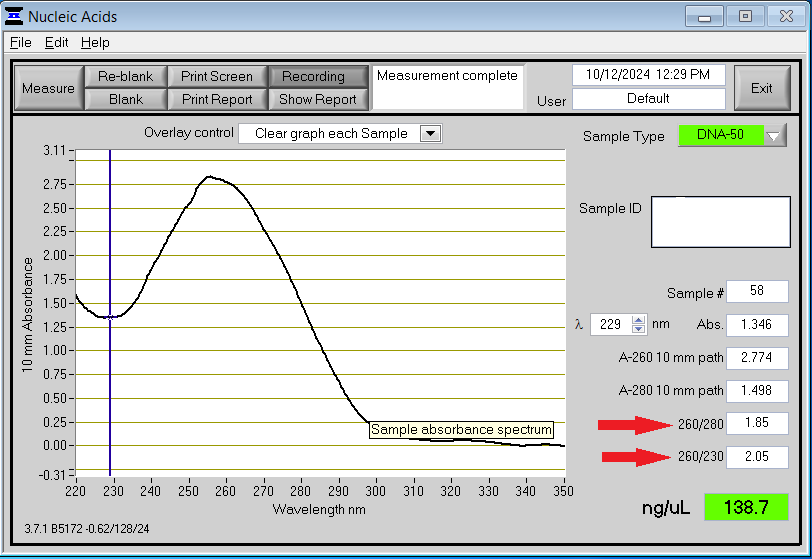
You will notice a strong peak at 260 nm, which is where genetic material is detected. It is important to look at the 260/280 and 260/230 ratios (red arrows). A good 260/280 ratio should be between 1.8 and 2.0 for DNA and between 2.0 and 2.2 for RNA. An ideal 260/230 ratio should be between 2.0 and 2.2. Below is a nanodrop result for a poor quality sample.
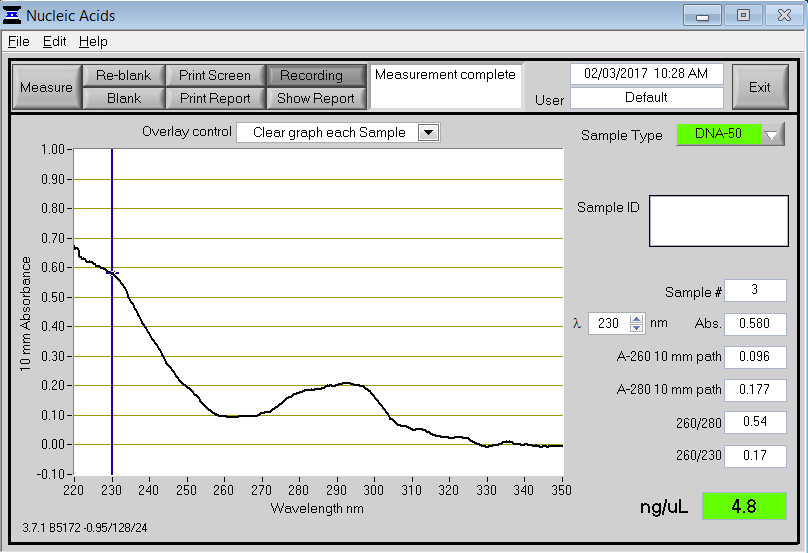
In this case, the curve looks much different without a peak at 260 nm. Both the 260/280 and 260/230 ratios are very low, indicating the presence of contaminants. A peak at 230 nm indicates the presence of organic compounds (e.g., Trizol and guanidine thiocyanate). A peak at or near 280 nm indicates contamination, typically from proteins or acidic phenol.
TapeStation
What analysis should I use?
The first thing to consider is what your sample type is: RNA, DNA or genomic material. For genomic DNA, please select the genomic DNA service.
For RNA and DNA there is a choice between high-sensitivity and not. High-sensitivity is only required when the sample concentration is too low to be run on the regular assay, typically lower than 25 ng/µL for the RNA assay and XX for DNA.
For DNA, product size also needs to be considered:
- If your product is under 1000 bp please select the D1000 assay (regular or high-sensitivity)
- If it is between 1000 and 5000 bp please select the D5000 assay (regular or high-sensitivity)
- For anything larger than 5000 bp please select the gDNA assay
On your order form please include which analysis should be done (DNA, genomic DNA, high-sensitivity DNA, RNA, high-sensitivity RNA).
If you are unsure about what assay to select please contact us to discuss your needs.
How should I provide my samples?
| # of Samples | Recommended storage tube/plate |
|---|---|
| 1 to 8 | Individual tubes |
| 8 to 24 | 8 tube strips or partial 96 well PCR plate* |
| More than 24 | Partial or whole 96 well PCR plate* |
*if using a partial 96-well plate please organize samples vertically (ie. A1 through H1, A2 through H2 etc) and not horizontally (ie. A1 through A12, B1 through B12).
Please label samples in individual tubes or strips clearly by number (1-96).
How much sample do I need to submit?
| Assay | Minimum Volume |
|---|---|
| RNA | 2 uL |
| HS RNA | 3 uL |
| DNA 1000 | 2 uL |
| HS DNA 1000 | 3 uL |
| DNA 5000 | 2 uL |
| HS DNA 5000 | 3 uL |
| gDNA | 2 uL |
What do my TapeStation results mean?
The TapeStation result will really depend on what kind of sample you are testing. It will vary significantly if it is extracted RNA, an amplified DNA product, a fragmented library prepared for NGS, or genomic DNA. Below are some examples of common sample types and what the results often look like. If you're ever unsure about what your results mean, please contact us.
Amplicon Samples
For amplicons, you should notice a fairly pronounced peak at an approximate size of the amplicon that you are trying to amplify using PCR (714 in the above example). You will also notice a "Lower" and "Upper" peak that are added to each sample to provide a frame of reference for the TapeStation to determine peak sizes of your sample.
Fragmented libraries ex. for NGS

A fragmented library, such as those used in Illumina NGS applications will have a more gradual peak, along with peaks for the "Lower" and "Upper" markers. The peak is more gradual in this case as the DNA is fragmented into varying lengths. In this case the peak size (440 in the above example) is more indicative of an average size of the fragmented DNA.
RNA samples

You will notice that for RNA samples from eukaryotic organisms, there are three peaks: the first is the "Lower" marker, one labelled 18S and the other 28S. The 18S and 28S peaks correspond to RNA from the ribosomal subunits. In between these peaks, there are areas indicating the presence of RNA from non-ribosomal subunits.
RNA samples also have one more consideration: the RNA Integrity Number or RIN. In the above example, you can see there is a value of 9.7 which is highlighted in green. The RIN indicates how degraded a sample is and ranges from 1 to 10, with 10 being RNA that has very little degradation and 1 being a very degraded sample. This value is determined based upon a ratio of the 18S and 28S subunits.
Genomic DNA

Genomic DNA TapeStation will typically have two peaks, one indicating the "Lower" marker and a peak at a size approximately the size of the genome in your sample.
Fragment Analysis
How should I provide my samples?
Fragment analysis samples should be provided in 96-well format.
What size fragments are detectable?
- 35–600 bp with LIZ 600
- 35–1,200 bp with LIZ 1200
