Protein ID info
Protein Identification Workflow
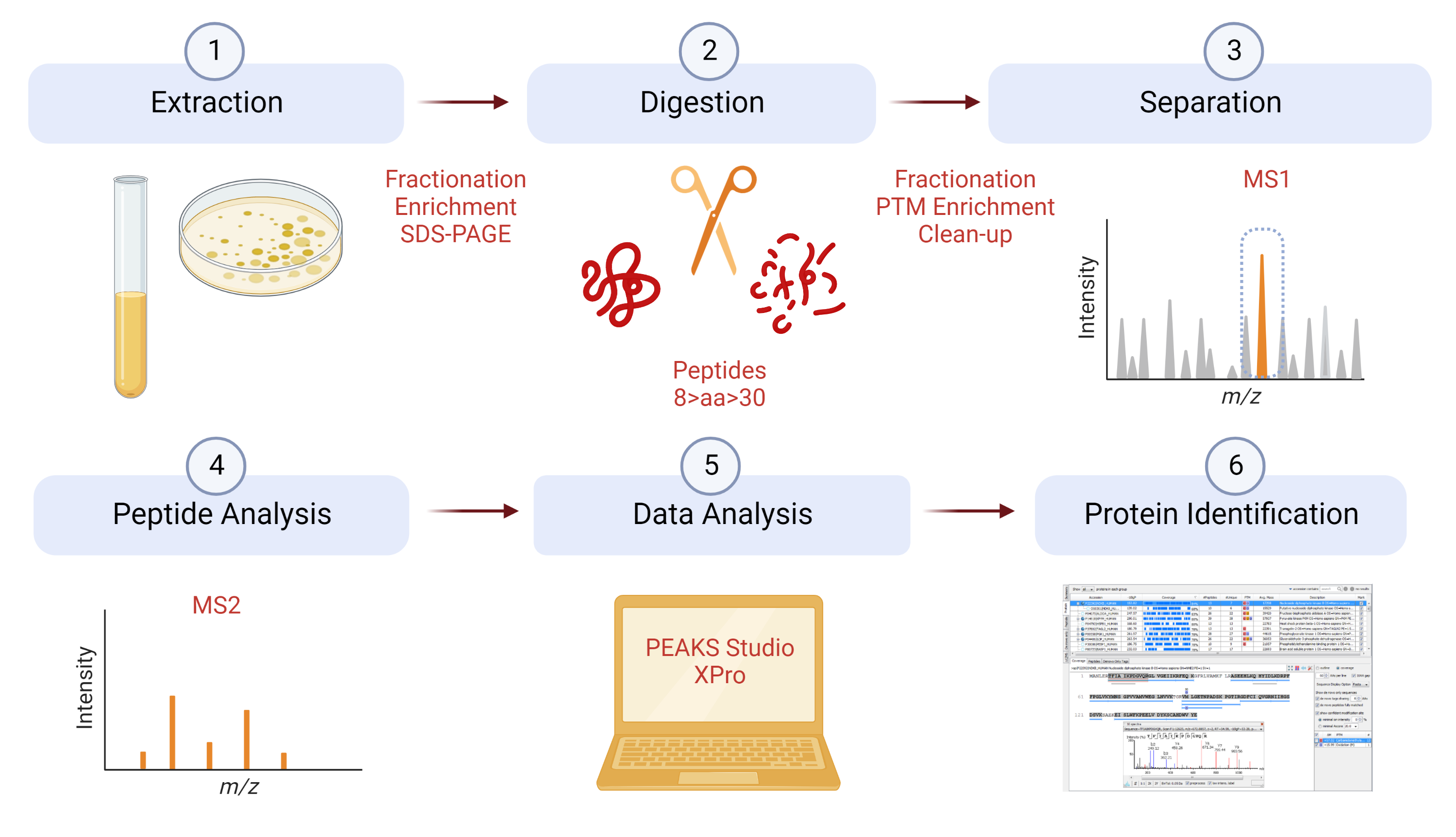
If you plan to submit any samples for protein identification, it is important that you read the information in this section, to save time and money by avoiding common mistakes.
- Sample preparation guidelines for protein ID [1]
- Co-IP: [2]Co-IP [3], MS compatible Flag tag Co-IP protocol [4], endogenous Co-IP [5],
- Protocol for solution digestion [6]
- Protocol for in-gel digestion [7]